Our scientists who created the assay explain the process.
The MSI-plus assay test uses multiplex PCR to amplify 14 highly sensitive mononucleotide repeats, the BRAF c.1799T>A allele, and mutation hotspots in KRAS and NRAS. Amplicons are sequenced followed by automated analysis to call MSI status and BRAF/RAS variants, providing data to interpret immune checkpoint inhibitor and anti-EGFR response as well as Lynch syndrome risk.
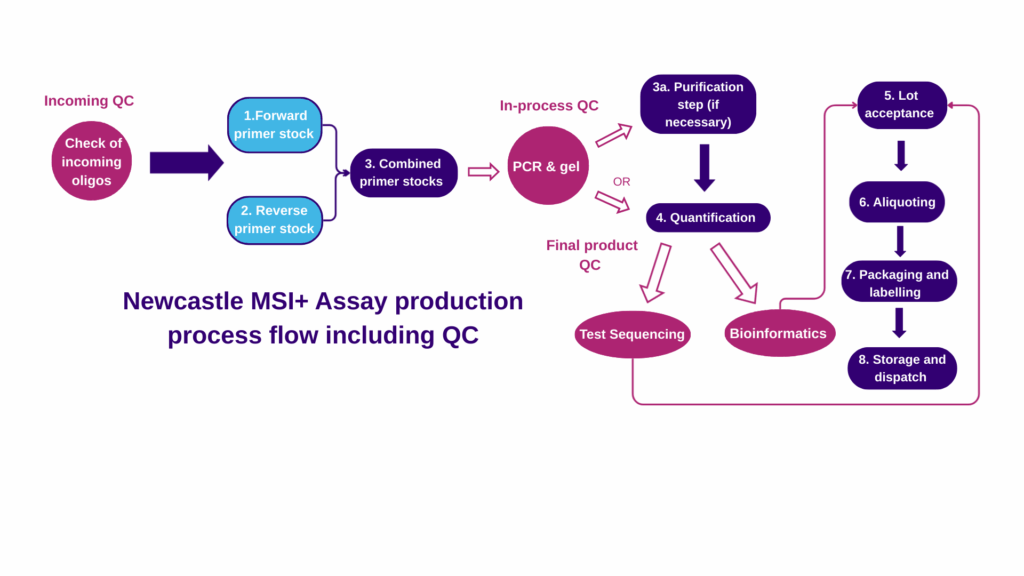
The MSI-plus assay test is quick and scalable, with batches of up to 96 samples being prepared for a single run. The assay has also been automated using the Tecan DreamPrep instrument providing an automatable workflow from PCR to NGS library pooling.

Each batch begins with a multiplex PCR that amplifies targets and incorporates Illumina adapters. It is robust to low quantity DNA, amplifying from <1ng of sample, and it has been validated FFPE curls. The mononucleotide repeats are monomorphic and so matched normal DNA is not required.
Amplicons are purified and pooled to make the NGS library. Sequencing on either Illumina MiSeq or NextSeq platforms is supported; validation on the NovaSeq is ongoing. The MSI-Plus amplicon library can be combined with other Nextera libraries using ≥100bp read lengths.
Sequence analysis uses a bespoke, automated pipeline consisting of read alignment (BWA) followed by variant calling and MSI score calculation (copyright R scripts). Results are under-pinned by quality control metrics to ensure reliability. The MSI-App summarises results and quality control metrics in a user-friendly and comprehensive output.
Please see the BJC Reports paper describing the MSI-Plus Assay development, validation, and clinical deployment
Make a purchase